Chapter 8 Profile Correlations
Next, we calculate profile correlations to test the stability of edges across all possible pairwise combinations of ages. A strong parallel correlation indicates that the weight of the edges is associated across two ages. We then map these profile correlations onto a heat map that displays the pairwise profile correlations. We would expect to see the strongest profile correlations cluster on the diagonal of heatmap – that is between ages that are adjacent or near in age.
## Multi-Trait
wide_fun <- function(df){
df <- unclass(df) %>% data.frame %>%
select(-from, -to) %>% spread(key = edge, value = weight)
rownames(df) <- df$age
df <- df %>% select(-age)
return(df)
}
# change the direction of the data
MT_procor <- MT_net_nested %>%
unnest(edges.df) %>%
group_by(inventory) %>%
nest() %>%
mutate(wide = map(data, wide_fun),
procor = map(wide, ~cor(t(.), use = "pairwise.complete.obs")))
# separate out differentiation from coherence
edges.mat50bw <- MT_net_nested %>%
filter(inventory == "IPIP50") %>% unnest(edges.df) %>%
separate(from, into = c("from_factor", "from_item"), 1) %>%
separate(to, into = c("to_factor", "to_item"), 1) %>%
filter(from_factor != to_factor) %>%
select(age, weight, edge) %>%
spread(key = edge, value = weight) %>%
unclass %>% data.frame
# separate out coherence from differentiation
edges.mat50wi <- MT_net_nested %>%
filter(inventory == "IPIP50") %>% unnest(edges.df) %>%
separate(from, into = c("from_factor", "from_item"), 1) %>%
separate(to, into = c("to_factor", "to_item"), 1) %>%
filter(from_factor == to_factor) %>%
select(age, weight, edge) %>%
spread(key = edge, value = weight) %>%
unclass %>% data.frame
# set the rownsames of these to age
rownames(edges.mat50bw) <- edges.mat50bw$age
rownames(edges.mat50wi) <- edges.mat50wi$age
##############################################
############ Profile Correlations ############
##############################################
# calculate profile correlations
procor50bw <- cor(t(edges.mat50bw[,-1]), use = "pairwise.complete.obs")
procor50wi <- cor(t(edges.mat50wi[,-1]), use = "pairwise.complete.obs")8.1 Profiles Correlation Plots (Figure 5)
##############################################
################## PLOTS #####################
##############################################
procor_plot_fun <- function(df, inv, leg){
df[upper.tri(df, diag = T)] <- NA
p <- tbl_df(df) %>%
mutate(age1 = colnames(.)) %>%
gather(key = age2, value = r, 1:(ncol(.) - 1)) %>%
filter(!is.na(r)) %>%
ggplot(aes(x = age2, y = age1, fill = r)) +
geom_raster(aes(fill = r)) +
scale_fill_gradient2(low = "blue", high = "red", mid = "white",
midpoint = .5, limit = c(0,1), space = "Lab",
name="Profile\nCorrelation") +
labs(x = "Age", y = "Age", title = sprintf("Profile Correlations for %s", inv)) +
theme_classic() +
theme(legend.position = leg,
axis.text = element_text(face = "bold"),
axis.text.x = element_text(angle = 90, size = rel(.8)),
plot.title = element_text(hjust = .5))
# ggsave(p, file = sprintf("%s/photos/Big5_%s_profile_cors_heatmap.png", data_path, inv), width = 12, height = 8)
p
}
MT_procor <- MT_procor %>%
mutate(plot = map2(procor, inventory, ~procor_plot_fun(.x, .y, leg = "none")))
layout <- rbind(c(1, 1, 2),
c(1, 1, 3))
p <- gridExtra::grid.arrange(
MT_procor$plot[[2]],
procor_plot_fun(procor50bw, "IPIP50 Cross-Trait Edges", leg = "none"),
procor_plot_fun(procor50wi, "IPIP50 Within-Trait Edges", leg = "none"),
layout_matrix = layout)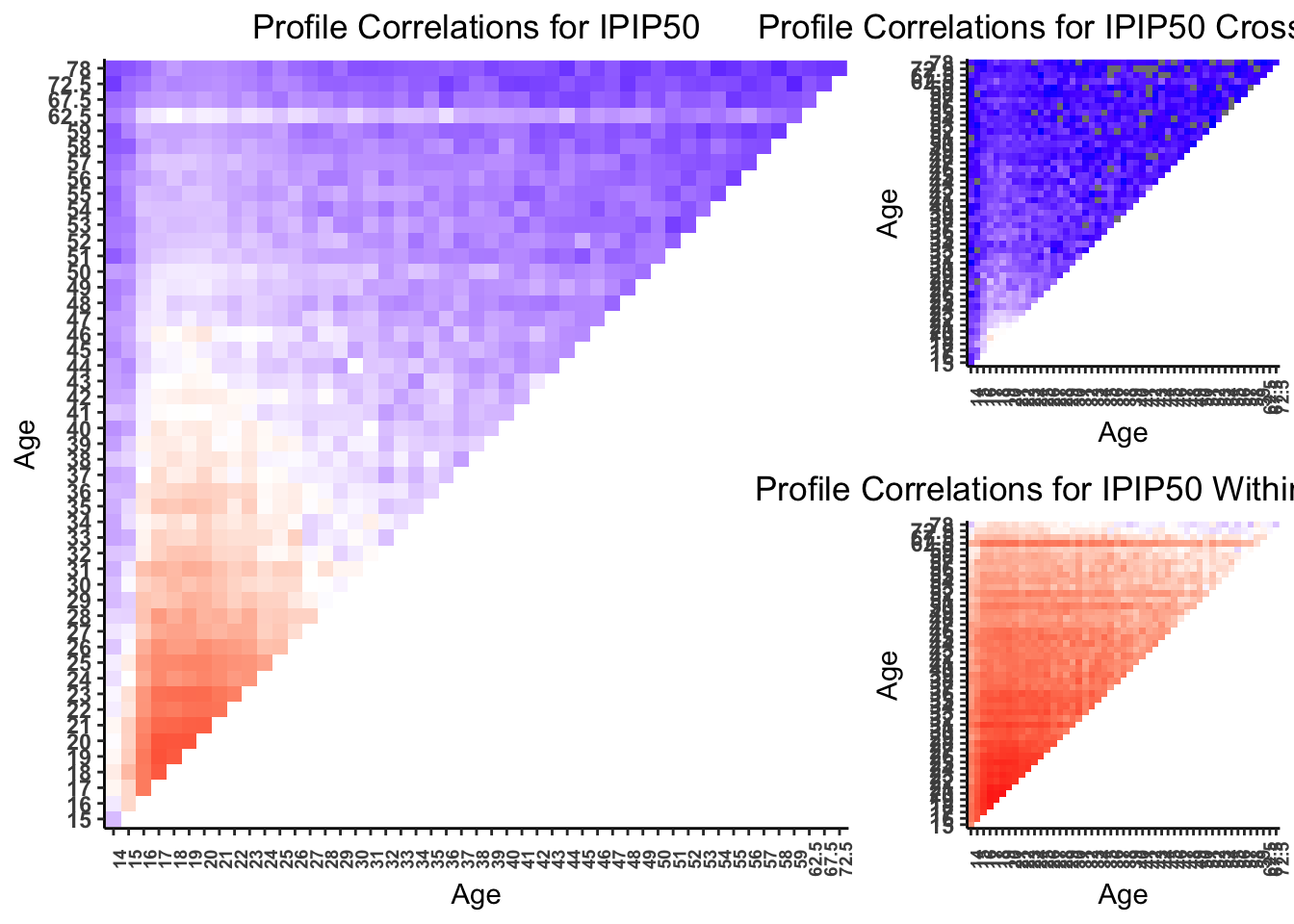
# ggsave(p, file = sprintf("%s/photos/Big5_IPIP50_profile_cors_heatmap.png", data_path),
# width = 12, height = 8)
rm(list = ls())